- Jing Y, Hu Z, Fan P, Xue Y, Wang L, Tarter RE, Kirisci L, Wang J, Vanyukov M, and Xie XQ. Analysis of substance use and its outcomes by machine learning I. Childhood evaluation of liability to substance use disorder. Drug Alcohol Depend. 2020 Jan 1; 206:107605. doi: 10.1016/j.drugalcdep.2019.107605. Epub 2019 Oct 22. PMID:31839402
- Hu Z, Jing Y, Xue Y, Fan P, Wang L, Vanyukov M, Kirisci L, Wang J, Tarter RE, and Xie XQ. Analysis of substance use and its outcomes by machine learning: II. Derivation and prediction of the trajectory of substance use severity. Drug Alcohol Depend. 2020 Jan 1;206:107604. doi: 10.1016/j.drugalcdep.2019.107604. Epub 2019 Oct 1. PMID:31615693
- Lee JY, Krieger JM, Li H, Bahar I. Pharmmaker: Pharmacophore modeling and hit identification based on druggability simulations. Protein Sci. 2020 Jan;29(1):76-86. doi: 10.1002/pro.3732. Epub 2019 Dec 4. PMID:31576621 PMCID:PMC6933858
- Zhang Y, Doruker P, Kaynak B, Zhang S, Krieger JM, Li H, Bahar I*. Intrinsic dynamics is evolutionarily optimized to enable allosteric behavior. Curr Opin Struct Biol. 2019 Nov 27;62:14-21. [Epub ahead of print]. doi: 10.1016/j.sbi.2019.11.002. PMID: 31785465
- Wang H, Wei Y, Cao M, Xu M, Wu W, & Xing EP. Deep inductive matrix completion for biomedical interaction prediction. IEEE International Conference on Bioinformatics and Biomedicine (IEEE BIBM 2019). This is a refereed conference paper/proceeding.
- Wu X, Mao Y, Wang H, Zeng X, Gao X, Xing EP, and Xu M., Regularized adversarial training (RAT) for robust cellular electron cryo tomograms classification. IEEE International Conference on Bioinformatics and Biomedicine (IEEE BIBM 2019). This is a refereed conference paper/proceeding.
- Wang Y, Guo H, Feng Z, Wang S, Wang Y, He Q, Li G, Lin W, Xie X-Q* and Lin Z*. PD-1-targeted discovery of peptide inhibitors by virtual screening, molecular dynamics simulation, and surface plasmon resonance. Molecules, 2019, 24(20), 3784; PMCID: PMC6833008; PMID: 31640203
- Zhang Y, Doruker P, Kaynak B, Zhang S, Krieger JM, Li H, Bahar I*. Intrinsic dynamics is evolutionarily optimized to enable allosteric behavior. Curr Opin Struct Biol. 2019 Nov 27;62:14-21. [Epub ahead of print]. doi: 10.1016/j.sbi.2019.11.002. PMID: 31785465
- Belovich AN, Aguilar JI, Mabry SJ, Cheng MH, Zanella D, Hamilton PJ, Stanislowski DJ, Shekar A, Foster JD, Bahar I, Matthies HJG, Galli A. A network of phosphatidylinositol (4,5)-bisphosphate (PIP2) binding sites on the dopamine transporter regulates amphetamine behavior in Drosophila Melanogaster. Mol. Psychiatry. 2019 Dec 3. [Epub ahead of print]. doi: 10.1038/s41380-019-0620-0. PMID:31796894
- Wang H, Lu C, Wu W & Xing EP. Graph-structured sparse mixed models for genetic association with confounding factors correction. IEEE International Conference on Bioinformatics and Biomedicine (IEEE BIBM 2019). This is a refereed conference paper/proceeding.
- Man VH*, He X, Ji B, Liu S, Xie XQ, and Wang J*. Molecular mechanism and kinetics of amyloid-β42 aggregate formation: A simulation study. ACS Chem Neurosci. 2019 Nov 20;10(11):4643-4658. Epub 2019 Nov 11. doi:10.1021/acschemneuro.9b00473. PMID:31660732
- Kim Y, Jun I, Shin DH, Yoon JG, Piao H, Jung J, Park HW, Cheng MH, Bahar I, Whitcomb DC, Lee MG. Regulation of CFTR Bicarbonate Channel Activity by WNK1: Implications for Pancreatitis and CFTR-Related Disorders. Cell Mol Gastroenterol Hepatol. 2020;9(1):79-103. doi: 10.1016/j.jcmgh.2019.09.003. Epub 2019 Sep 24. PMID:31561038
- Liu S, He X, Man VH, Ji B, Liu J*, and Wang J*. New application of in silico methods in identifying mechanisms of action and key components of anti-cancer herbal formulation YIV-906 (PHY906). Phys Chem Chem Phys 2019 Nov 14; 21(42):23501-23513. Epub 2019 Oct 16. doi: 10.1039/c9cp03803e. PMID:31617551
- Wang H, Yue T, Yang J, Wu W and Xing, EP*., 2019. Deep mixed model for marginal epistasis detection and population stratification correction in genome-wide association studies. BMC Bioinformatics, 20 (Suppl 23), pp.1-11, PMCID: PMC6933893.
- Cheng J, Wang S, Lin W, Wu N, Wang Y, Xie X-Q*, and Feng Z*. Computational System Pharmacology-Target Mapping for Fentanyl-laced Cocaine Overdose. ACS Chem. Neuro. 2019 Jul 15. doi: 10.1021/acschemneuro.9b00109. PMID: 31257858.
- Ma S, Attarwala I, Xie X-Q. SQSTM1/p62: a potential target for neurodegenerative disease. ACS Chem Neurosci 2019 Jan 18. doi: 10.1021/acschemneuro.8b00516.
- Wu N, Feng Z, He X, Kwon W, Wang J*, Xie X-Q*. Insight of Captagon Abuse by Chemogenomics Knowledgebase-guided Systems Pharmacology Target Mapping Analyses. Sci Rep 2019 Feb 19;9(1):2268: PMID: 30783122 PMCID: PMC6381188.
- Cheng MH, Bahar I. Monoamine transporters: structure, intrinsic dynamics and allosteric regulation, Nature Structural & Molecular Biology 2019; 26: 545–556.
- Taylor DL, Gough A, Schurdak ME, Vernetti L, Chennubhotla CS, Lefever D, Pei F, Faeder JR, Lezon TR, Stern AM, Bahar I. “Harnessing Human Microphysiology Systems as Key Experimental Models for Quantitative Systems Pharmacology” in Handbook of Experimental Pharmacology, 2019, p 1-41.
- Bian YM, He XB, Jing YK, Wang LR, Wang JM, Xie XQ. Computational systems pharmacology analysis of cannabidiol: a combination of chemogenomics-knowledgebase network analysis and integrated in silico modeling and simulation. Acta Pharmacologica Sinica. 2019 Mar;40(3):374. PMID: 30202014 PMCID: PMC6460368.
- Cheng MH, Ponzoni L, Sorkina T, Lee JY, Zhang S, Sorkin A, Bahar I. Trimerization of Dopamine Transporter Triggered by AIM-100 Binding: Molecular Mechanisms and Effect of Mutations. Neuropharmacology 2019 [Epub ahead of print]
- Zhou Z, Feng Z, Hu D, Yang P, Gur M, Bahar I, Cristofanilli M, Gradishar WJ, Xie XQ, Wan Y. A novel small-molecule antagonizes PRMT5-mediated KLF4 methylation for targeted therapy. EBioMedicine 2019; 19: 30312-30313. PMID: 31101597
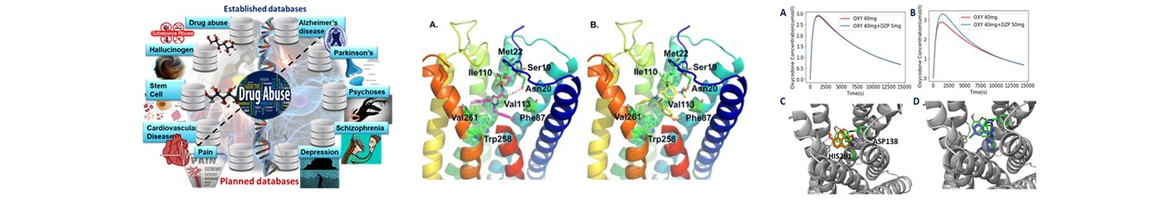
- Pei F, Li H, Liu, B, Bahar I. Quantitative systems pharmacological analysis of drugs of abuse reveals the pleiotropy of their targets and the effector role of mTORC1. Front. in Pharm. 2019 [Epub ahead of print] PMID: 30906261
- Mikulska-Ruminska K, Shrivastava IH, Krieger JM, Zhang S, Li H, Bayir H, Wenzel SE, VanDemark AP, Kagan VE, Bahar I. Characterization of differential dynamics, specificity, and allostery of lipoxygenase family members. J Chem Inf Model. 2019 [Epub ahead of print] PMID: 30762363
- Wang Y, Lin W, Wu N, Wang S, Chen M, Lin Z, Xie X-Q*, and Zhi-wei Feng*. Structural insight into the serotonin (5-HT) receptor family by molecular docking, molecular dynamics simulation and systems pharmacology analysis. Acta Pharmacologica Sinica, 2019, in press.
- Zhang S, Li H, Krieger JM, Bahar I. Shared signature dynamics tempered by local fluctuations enables fold adaptability and specificity. Mol Biol & Evol 2019 [Epub ahead of print] PMID: 31028708
- Bian Y, Jing Y, Wang L, Ma S, Jun JJ, Xie XQ. Prediction of Orthosteric and Allosteric Regulations on Cannabinoid Receptors Using Supervised Machine Learning Classifiers. Mol Pharm. 2019 Jun 3;16(6):2605-2615. doi: 10.1021/acs.molpharmaceut.9b00182. Epub 2019 May 3. PMID: 31013097.
- Man, V.; Truong, P.; Li, M.; Wang, J.; Van-Oanh, N.-T.; Derreumaux, P.; Nguyen, P. Molecular Mechanism of the Cell Membrane Pore Formation Induced by Bubble Stable Cavitation, J. Phys. Chem. B, 2019, 123, 71-78
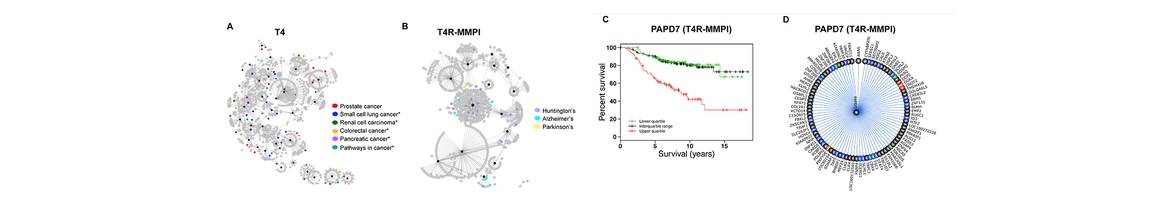
- Chen M, Jing Y, Wang L, Feng Z*, and Xie X-Q*. DAKB-GPCRs: An Integrated Computational Platform for Drug Abuse Related GPCRs. Journal of Chemical Information and Modeling, 2019, 59 (4), pp 1283–1289. DOI: 10.1021/acs.jcim.8b00623. PMID: 30835466.
- Fan P, Wang N, Wang L*, and Xie XQ*. Autophagy and apoptosis specific knowledgebases-guided systems pharmacology drug research. Current Cancer Drug Targets 2019 Feb 6;doi:10.2174/1568009619666190206122149. [Epub ahead of print]. PMID:30727895
- Wang H, Liu X, Tao Y, Ye W, Jin Q, Cohen JW, and Xing EP*. Automatic human-like mining and constructing reliable genetic association database with deep reinforcement learning, Proceedings of 24th Pacific Symposium on Biocomputing (PSB 2019). Publication is not a journal.
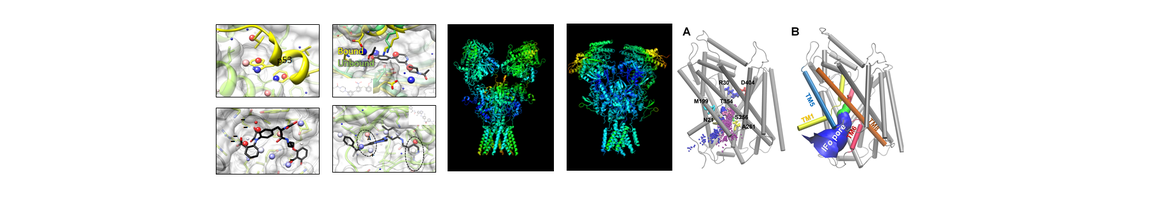
- Lee JY, Krieger J, Herguedas B, García-Nafría J, Dutta A, Shaikh SA, Greger IH, Bahar I. Druggability Simulations and X-ray Crystallography Reveal a Ligand-binding Site in the GluA3 AMPA Receptor N-terminal Domain. Structure 2019; 27: 241-252, PMID: 30528594
- Wang H, Wu Z, and Xing EP*. Removing confounding factors associated weights in deep neural networks improves the prediction accuracy for healthcare applications, Proceedings of 24th Pacific Symposium on Biocomputing (PSB 2019). Publication is not a journal.
- Ge, H. X.; Bian, Y. M.; He, H. X.; Xie, X. Q.; Wang, J. M., Significantly different effects of tetrahydroberberrubine enantiomers on dopamine D1/D2 receptors revealed by experimental study and integrated in silico simulation. J. Computer-Aided Molecular Design 2019, 33, 447-459. PMID: 30840169
- Wodak SJ, Paci E, Dokholyan NV, Berezovsky IN, Horovitz A, Li J, Hilser VJ, Bahar I, Karanicolas J, Stock G, Hamm P. Allostery in its many disguises: from theory to applications. Structure. 2019 Apr 2;27(4):566-578. doi: 10.1016/j.str.2019.01.003. Epub 2019 Feb 7. PMID: 30744993 PMCID: PMC6688844
- Wang L, Ma S, Hu Z, McGuire TF, Xie XQ. Chemogenomics Systems Pharmacology Mapping of Potential Drug Targets for Treatment of Traumatic Brain Injury. Journal of neurotrauma. 2019 Feb 1;36(4):565-75. PMID: 30014763 PMCID: PMC6354609 [Available on 2020-02-15] DOI: 10.1089/neu.2018.5757.
- Man VH, Li MS, Wang J, Derreumaux P, Nguyen PH. Interaction mechanism between the focused ultrasound and lipid membrane at the molecular level.J Chem Phys. 2019 Jun 7;150(21):215101. doi: 10.1063/1.5099008. PMID: 31176320
- Man VH, Li MS, Wang J, Derreumaux P, Nguyen PH. Nonequilibrium atomistic molecular dynamics simulation of tubular nanomotor propelled by bubble propulsion. The Journal of chemical physics. 2019 Jul 14;151(2):024103. PMID: 31301696 PMCID: PMC6624022
- Marchetti-Bowick M, Yu Y, Wu W, Xing EP. A penalized regression model for the joint estimation of eQTL associations and gene network structure. The Annals of Applied Statistics. 2019;13(1):248-70.