PI: Dr. Ivet Bahar
Dynamics of Neurotransmitter Transporters and Modulation of their Function (Dr. Mary Hongying Cheng)
Neurotransmitter: sodium symporters (NSSs) play a central role in neurotransmission. The function of NSSs is modulated by regulatory proteins, addictive drugs (e.g. cocaine and amphetamine) and lipids. We are particularly interested in developing and implementing multi-scale computing technology for investigating biological systems at both molecular-to-cellular levels and simulating spatio-temporal events relevant to biological function and dysfunction.
Our current research focuses on understanding the molecular mechanisms of i) transporter function (See Cheng and Bahar 2015; Cheng and Bahar 2014; Cheng et al 2017); ii) modulation of the transporter function by addictive drugs (i.e. amphetamine and cocaine; Cheng et al 2015; Ma et al 2017), and regulatory proteins (i.e. G-protein, and CaMKII); iii) mechanisms of NSS oligomerization and its potential allosteric modulation of the transporter function (Cheng et al 2017 and Gur et al 2017); and iv) transporter interactions with neuronal lipids (i.e., cholesterol and PIP2).
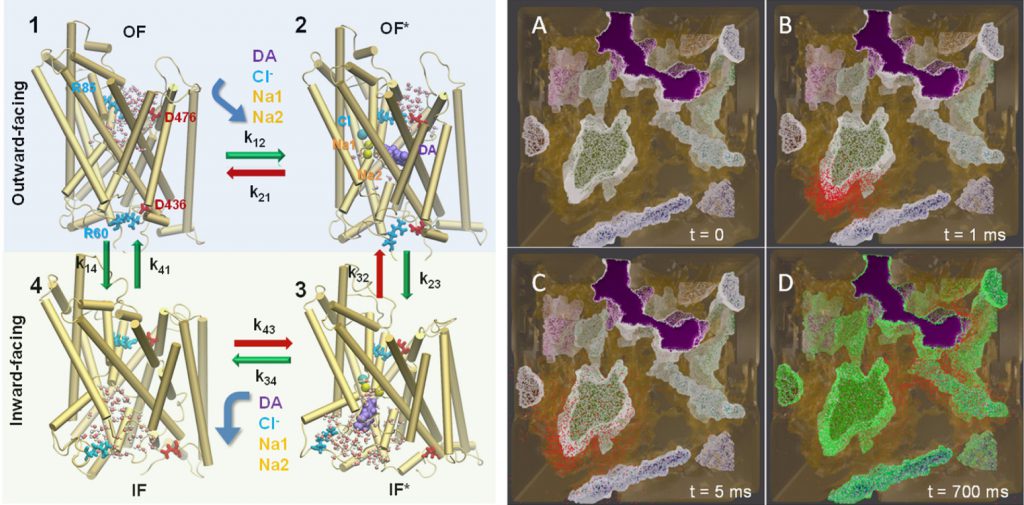
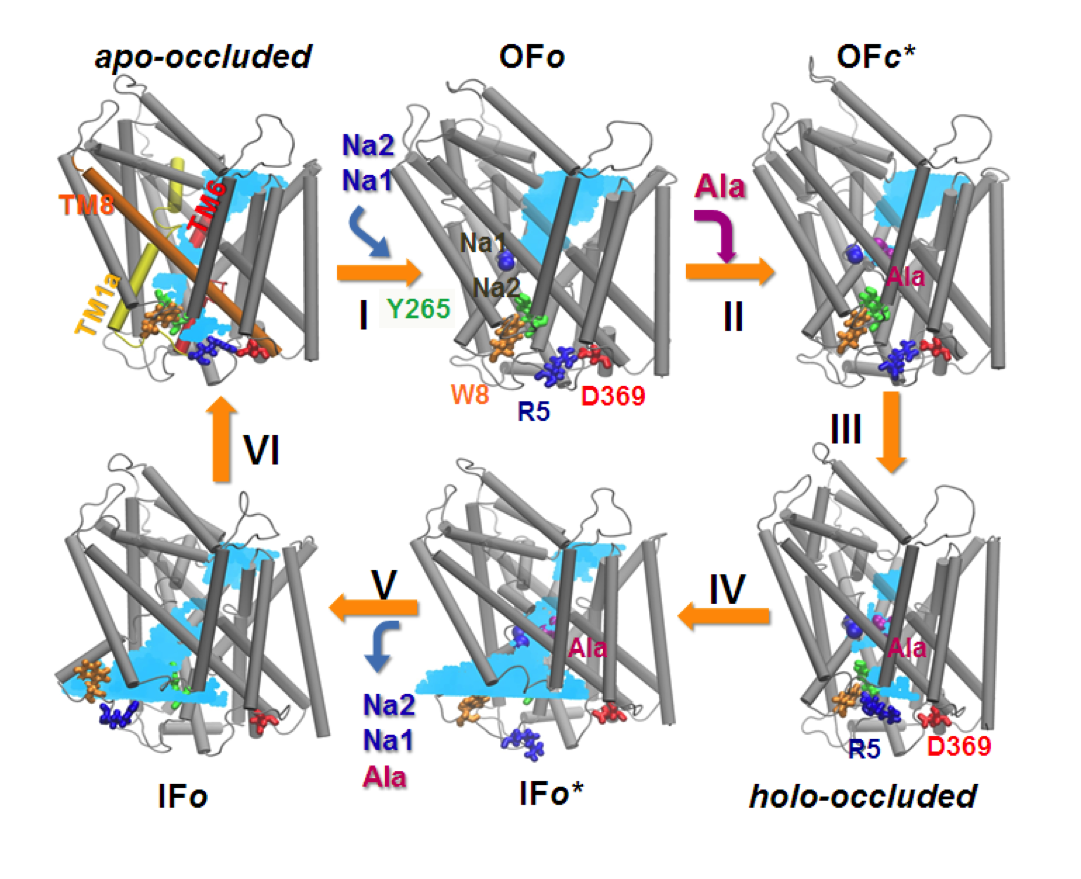 Conformational states visited during LeuT transport cycle and corresponding hydration patterns and changes in interactions at intracelluar and extracelluar gates. Six states, labeled, are distinguished, including three newly determined ones by our advanced MD simulations: holo-occluded, inward-facing substrate-bound open (IFo*), and apo-occluded. The putative two intracelluar gating pairs R5-D369 and W8-Y268-Y265 are detected. Hydrated regions are indicated by blue shaded areas. (Cheng and Bahar, 2014, PLos Comp. Biol.)
Conformational states visited during LeuT transport cycle and corresponding hydration patterns and changes in interactions at intracelluar and extracelluar gates. Six states, labeled, are distinguished, including three newly determined ones by our advanced MD simulations: holo-occluded, inward-facing substrate-bound open (IFo*), and apo-occluded. The putative two intracelluar gating pairs R5-D369 and W8-Y268-Y265 are detected. Hydrated regions are indicated by blue shaded areas. (Cheng and Bahar, 2014, PLos Comp. Biol.)
Cell Regulation Processes and Response to Dopamine Influx (Dr. Bing Liu)
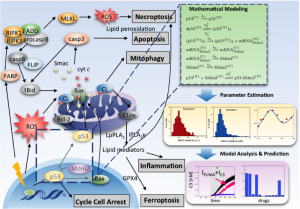
Drug abuse and related diseases influence the regulation of cellular processes such as cell survival/death and immune response. We are using systems biology approaches to understand the dynamics of the protein-protein interaction and metabolic networks and that underlies these cellular processes, and to predict (poly)pharmacological strategies for improving treatments. We have constructed kinetic models for a variety of cellular processes, including apoptosis (Liu et al, 2014), ferroptosis (Kagan et al, 2017), innate immune response (Liu et al, 2016), autophagy (Liu et al, 2017), and dopamine transporter trafficking and efflux. We are currently using these models to study the methamphetamine-induced autophagy-apoptosis cell decision and the amphetamine-induced regulation of dopamine transporter (in collaboration with the Amara lab at the NIMH, NIH), and the type-2 inflammation in asthma induced autophagic cell survival and ferroptotic cell death (in collaboration with the FRP PI Wenzel, Director of Asthma Institute at UPMC).
Stimulation and Allosteric Modulation of Receptors Involved in Regulating Neuronal and Immune System Signals (Drs. JiYoung Lee, James Krieger and HongChun Li)
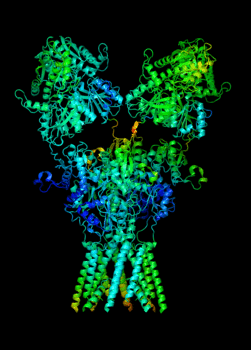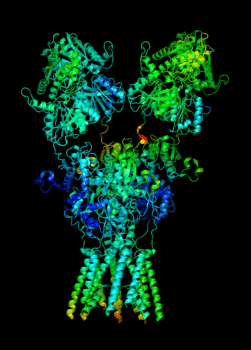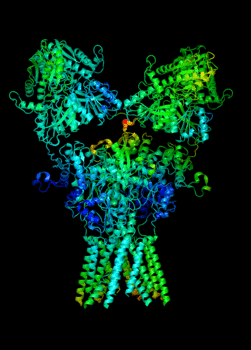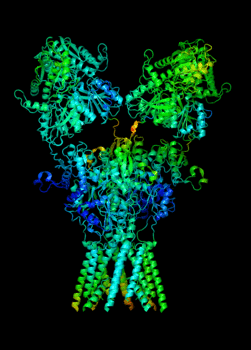
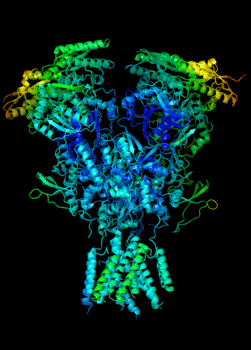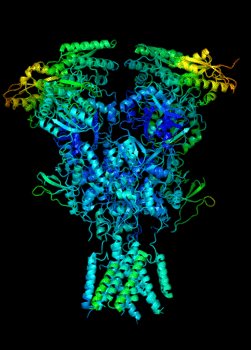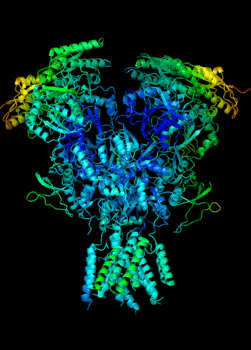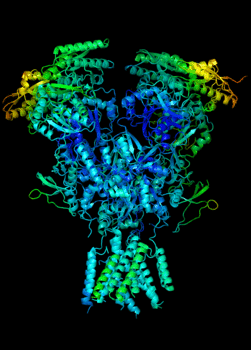 Elucidating the allosteric dynamics of receptors is very important for developing strategies toward modulating their activities. A major challenge is receptor structural flexibility, which mediates ligand binding, activation/inactivation/desensitization mechanisms as well as allosteric signaling events. This is a difficult problem because of the complexity and specificity of interactions, the multiplicity of conformations, and insufficient data on the structure and energetics of protein-ligand interactions. We are tackling this problem using network models for complex structures and molecular dynamics simulations at atomic level. An important class of targets playing a key role in synaptic plasticity and neurological disorders is the family of ionotropic glutamate receptors (iGluRs), iGluRs are ligand-gated ion channels and receptors at the same time, and they regulate excitatory neurotransmission. Using Anisotropic Network Model (ANM), we have recently shown how two prominent subfamilies of iGluR family, AMPA receptor (AMPAR) and NMDA receptor (NMDAR), share robust mechanisms of regulation, and could serve as targets for allosteric modulation of their activities. (see Dutta et al 2015; see more movies of global dynamics of AMPAR and NMDAR; see Krieger et al 2015). This was a collaborative study with the lab of Dr Ingo Greger at the MRC, Cambridge U, UK.
Elucidating the allosteric dynamics of receptors is very important for developing strategies toward modulating their activities. A major challenge is receptor structural flexibility, which mediates ligand binding, activation/inactivation/desensitization mechanisms as well as allosteric signaling events. This is a difficult problem because of the complexity and specificity of interactions, the multiplicity of conformations, and insufficient data on the structure and energetics of protein-ligand interactions. We are tackling this problem using network models for complex structures and molecular dynamics simulations at atomic level. An important class of targets playing a key role in synaptic plasticity and neurological disorders is the family of ionotropic glutamate receptors (iGluRs), iGluRs are ligand-gated ion channels and receptors at the same time, and they regulate excitatory neurotransmission. Using Anisotropic Network Model (ANM), we have recently shown how two prominent subfamilies of iGluR family, AMPA receptor (AMPAR) and NMDA receptor (NMDAR), share robust mechanisms of regulation, and could serve as targets for allosteric modulation of their activities. (see Dutta et al 2015; see more movies of global dynamics of AMPAR and NMDAR; see Krieger et al 2015). This was a collaborative study with the lab of Dr Ingo Greger at the MRC, Cambridge U, UK.
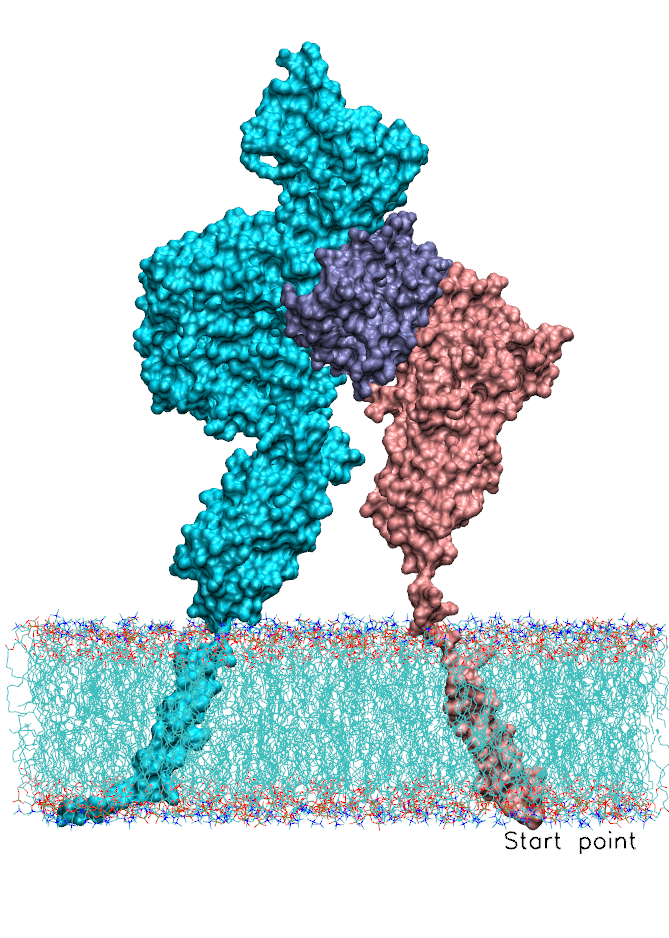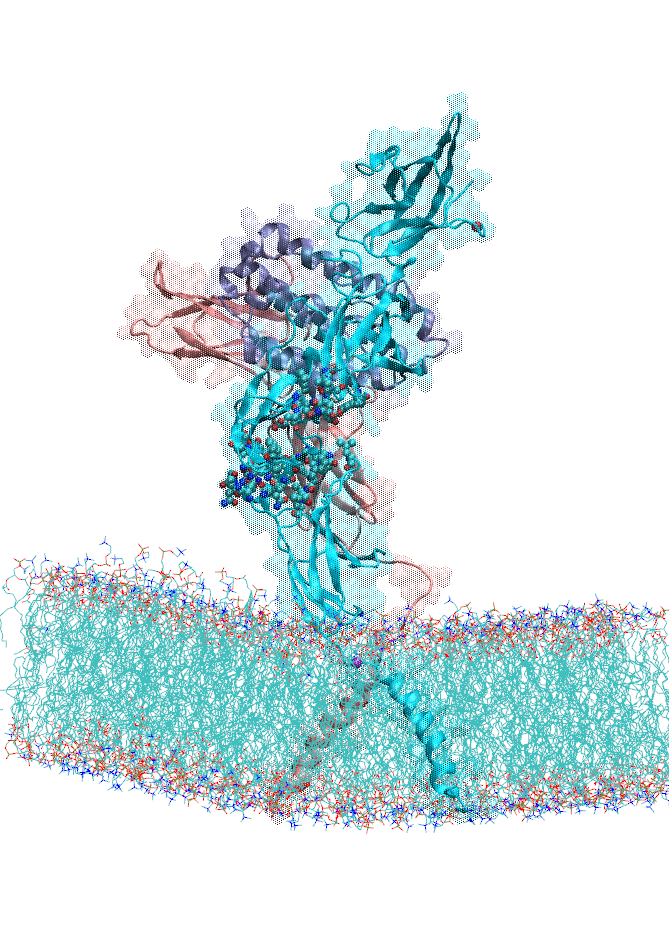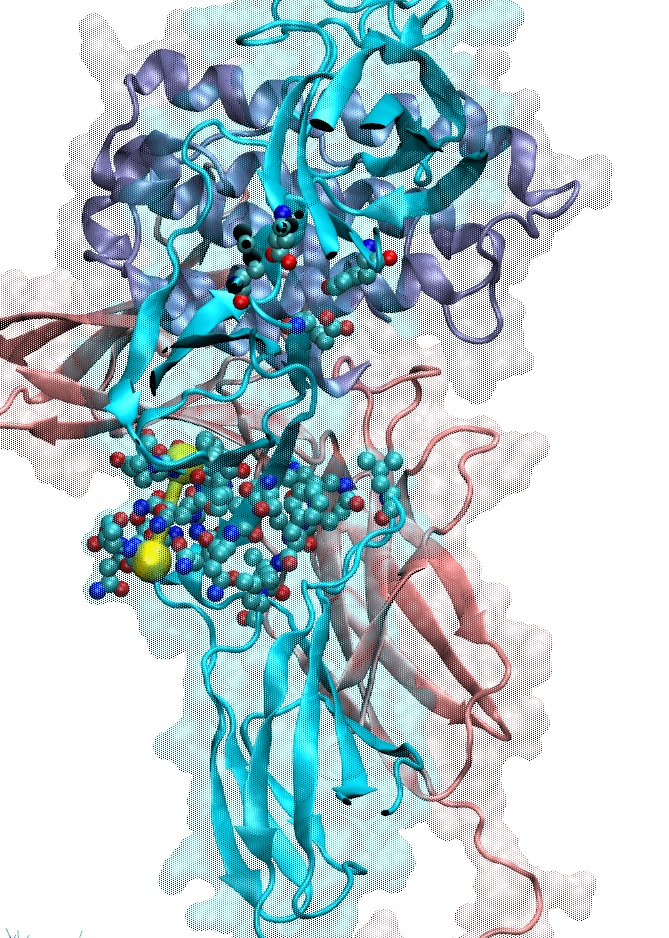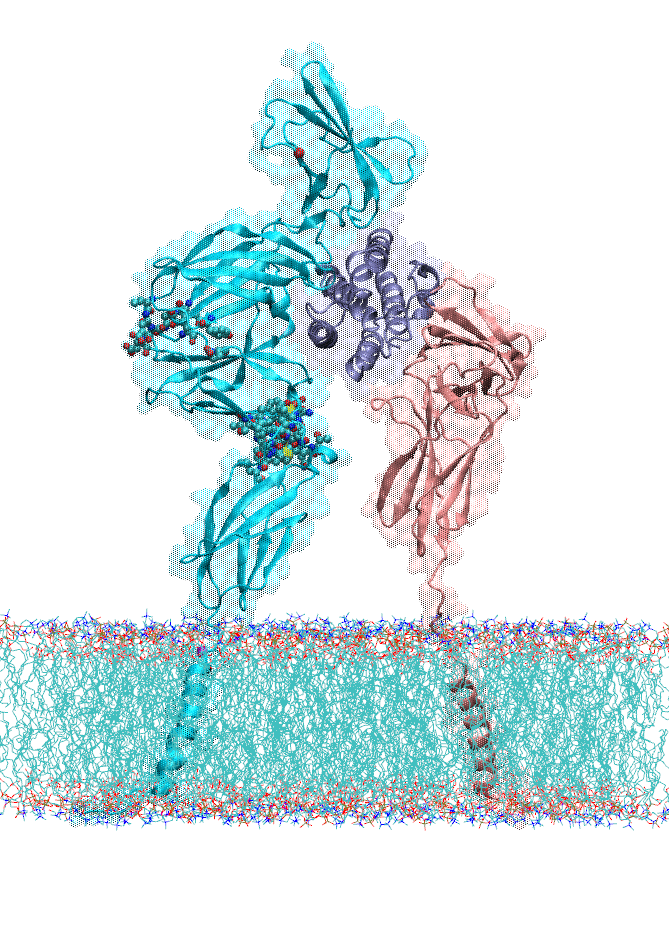
We also focused on interferon (IFN) signaling. IFNα2, a Type-I interferon, forms a ternary complex with two receptors, IFNAR1 and IFNAR2. We have shown that the binding affinity of IFNα2 to these receptors, as well as downstream signaling strength, can be modulated by altering the dynamics of the IFNAR1 (Li et at, 2017a). The global motions (see animation) of the IFNAR1-IFNα2-IFNAR2 ternary complex were predicted using the membrANM in which the lipid environment is included in the model and simulations. IFNα2 associated immune responses were shown to be modulated by introducing Cys-Cys double mutants which form cross-links between different subdomains of IFNAR1. This was a collaborative study with the Schreiber lab at the Weizmann Institute in Israel where experiments (binding and functional assays) have been performed.
Development of Server and Database for Applications of Elastic Network Models (Dr. HongChun Li)
The elastic network models (ENMs) are widely used in studying of the dynamics of biomolecules and identifying the modes of motions that are likely to be relevant to their biological functions. We have analyzed to date more than 90% PDB (Protein Data Bank) structures using the Gaussian Network Model (GNM) introduced by the Bahar lab. The pre-calculated equilibrium dynamics have been deposited in the iGNM 2.0 database (Li et at, 2016). We also integrated both the Gaussian Network Model (GNM) and the Anisotropic Network Model (ANM) in a newly designed DynOmics ENM server with user-friendly interfaces. The unique features of DynOmics include the inclusion of the effect of environment on collective dynamics, the prediction of potential functional sites implicated in biological activities and the reconstruction of all-atom conformers sampled along the dominant modes of motions intrinsically favored by the biomolecular architecture (Li et at, 2017b).
The effect of spatial complexity and heterogeneities on the efficiency of dopamine reuptake (Cihan Kaya)
Efficient clearance of neurotransmitters from the synapse by dopamine transporters (DATs) is critical to regulating dopamine (DA) signaling in the central nervous system. Despite significant advances in the field, we still lack a complete mechanistic understanding of DA transport events. We adopted a multiscale methodology to examine the effects of spatial complexity and firing patterns on DA reuptake by DATs: We used a kinetic scheme from one of our previous studies as input in MCell simulations together with fluorescence images of dopaminergic neurons (Block et al, 2015). DAT spatial distributions and structural heterogeneities were observed to alter the efficiency of DA reuptake, suggesting that realistic spatial descriptions are required to accurately assess the mechanism of DAT function. Using this model we also explored the effect of neurodegeneration on DA reuptake.
(Left panel) Schematic representation of the sequence of events occurring during the transport cycle of DAT. (Right panel) Snapshots from MCell simulations of DA release and reuptake by DATs on DA terminals.
Molecular Mechanism of Dopamine Transport by Human Dopamine Transporter
(Mary Hongying Cheng and Ivet Bahar Structure (2015))
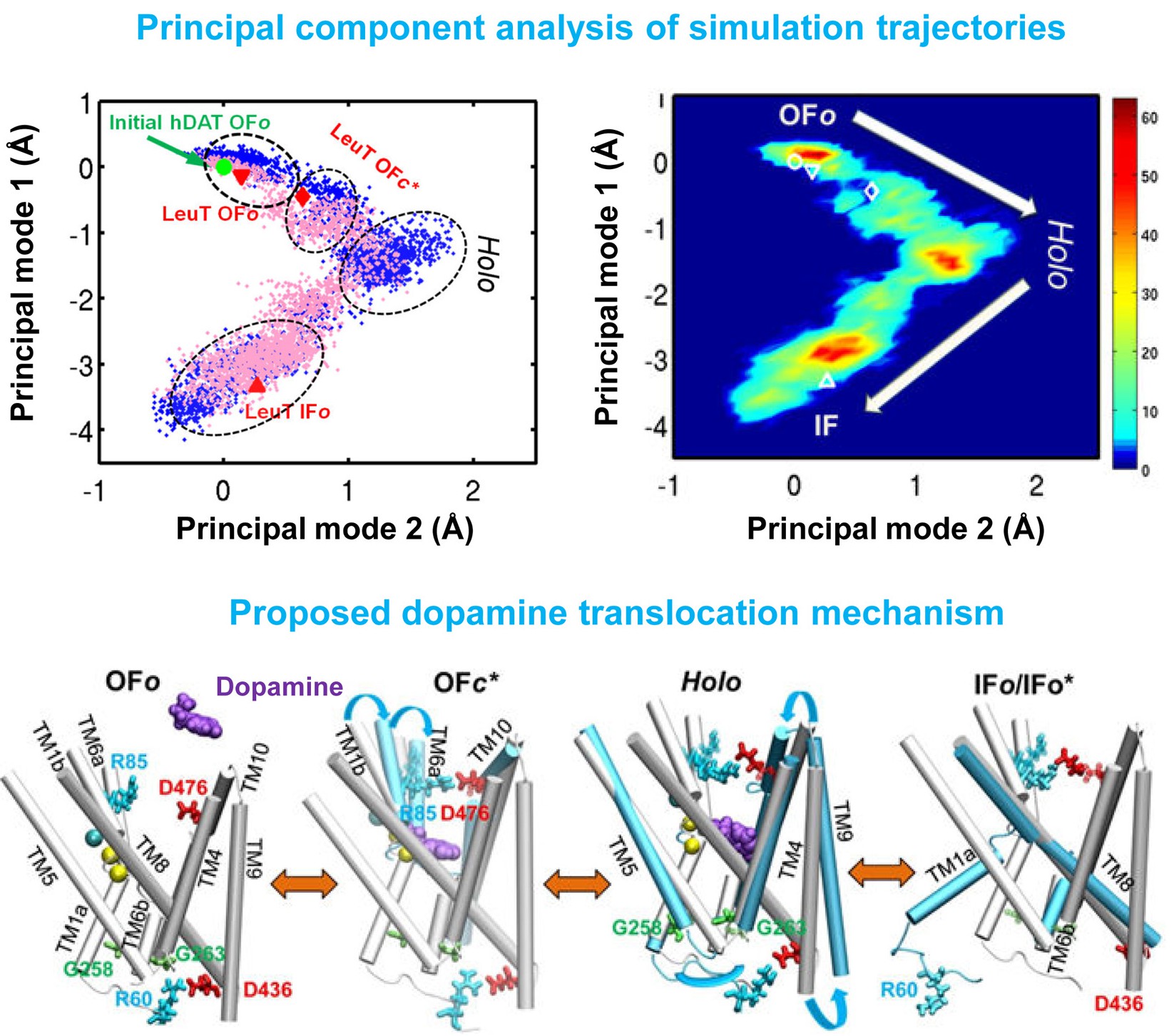
Dopamine transporter (DAT) controls neurotransmitter dopamine (DA) homeostasis by reuptake of excess DA from the synapse into the presynaptic neuron, assisted by the co-transport of two sodium ions. Malfunction of human DAT (hDAT) has been implicated in many neurological disorders. The first DAT structure (dDAT, from Drosophila melanogaster) has been recently resolved, which permit us to conduct a structure-based computational study of the time-resolved mechanism of DA reuptake by hDAT at full atomic scale. Using homology modeling and full-atomic microseconds accelerated simulation, we investigated the complete DA translocation including uptake from the EC region to its intracellular release and highlighted the key interactions that mediate DA translocation through hDAT. Our major observations are: spontaneous closure of extracellular gates prompted by DA binding; stabilization of a holo-occluded intermediate distinguished by close association of TM1b and TM6a with TM10 and resulting cluster of hydrophobic residues that prevents the hydration of the binding site; subsequent exposure to intracellular water triggered by Na2 dislocation, accompanied by a redistribution of salt bridges at the cytosolic surface; concerted tilting of TM5 and TM7, critical to opening the exit pore for substrate/ion channeling, enabled by the stretching of the G258-G263 loop, disruption of N82-N353 hydrogen bond, which drives the release of a Na+ ion and the Cl- ion; and DA release induced only after protonation of D79. Following Figure illustrates the transport mechanism, deduced from our over 13 sets micro-seconds MD simulations.
In the movie, purple balls represent dopamine molecule; yellow and cyan spheres the co-transported sodium and chloride. The residues forming the extracellular gate R85-D476 and intracellular gate R60-D436 are shown in cyan (basic) and red (acidic) sticks, respectively.
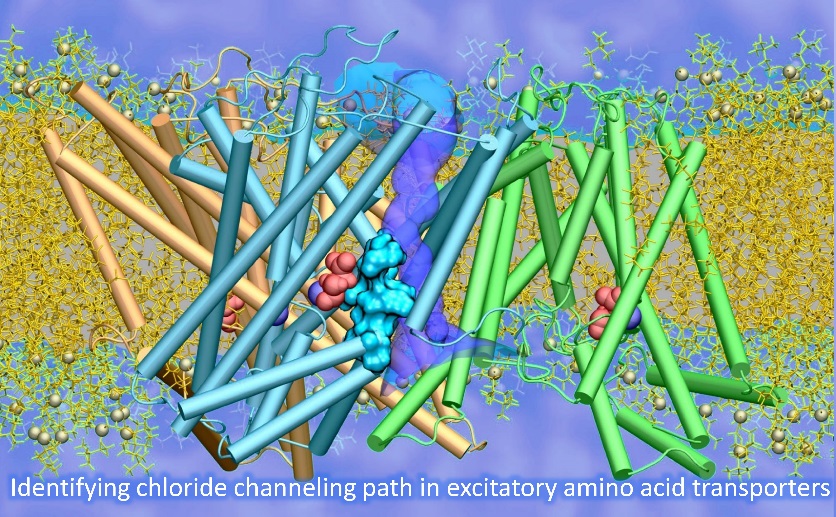
Substrate transport and anion permeation proceed through distinct pathways in glutamate transporters.
(Cheng MH*, Torres-Salazar D*, Gonzalez-Suarez AD, Amara SG, Bahar I. ELife 2017)
The paper provides novel evidence on the molecular determinants of the dual function, substrate transport and anion permeation, of excitatory amino acid transporters (EAATs). It opens avenues toward illuminating how EAATs regulate synaptic transmission and neurological conditions. This is a collaborative study between the Bahar and Amara (NIH) labs.