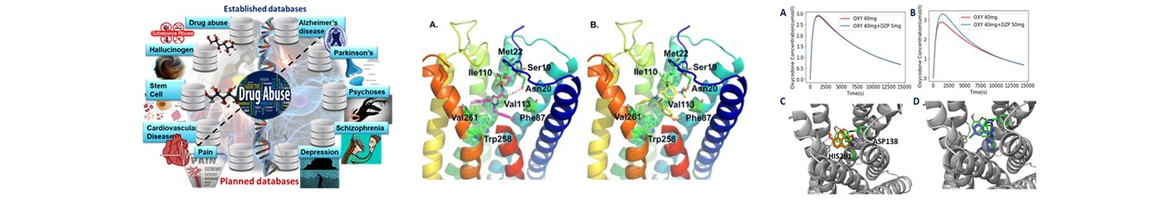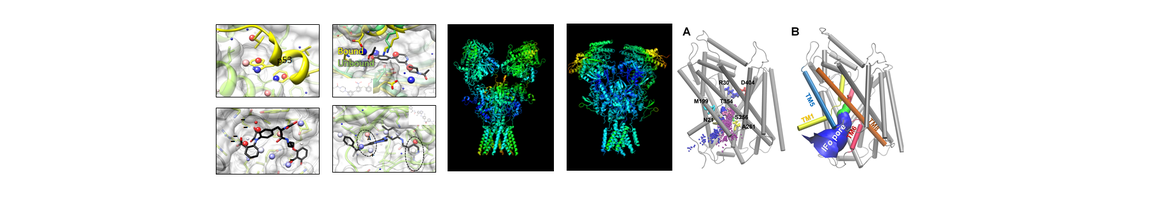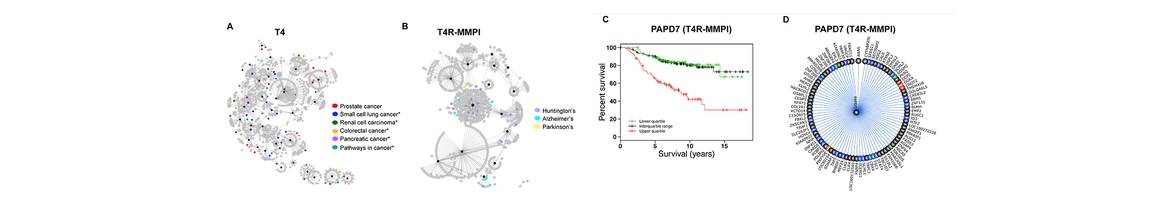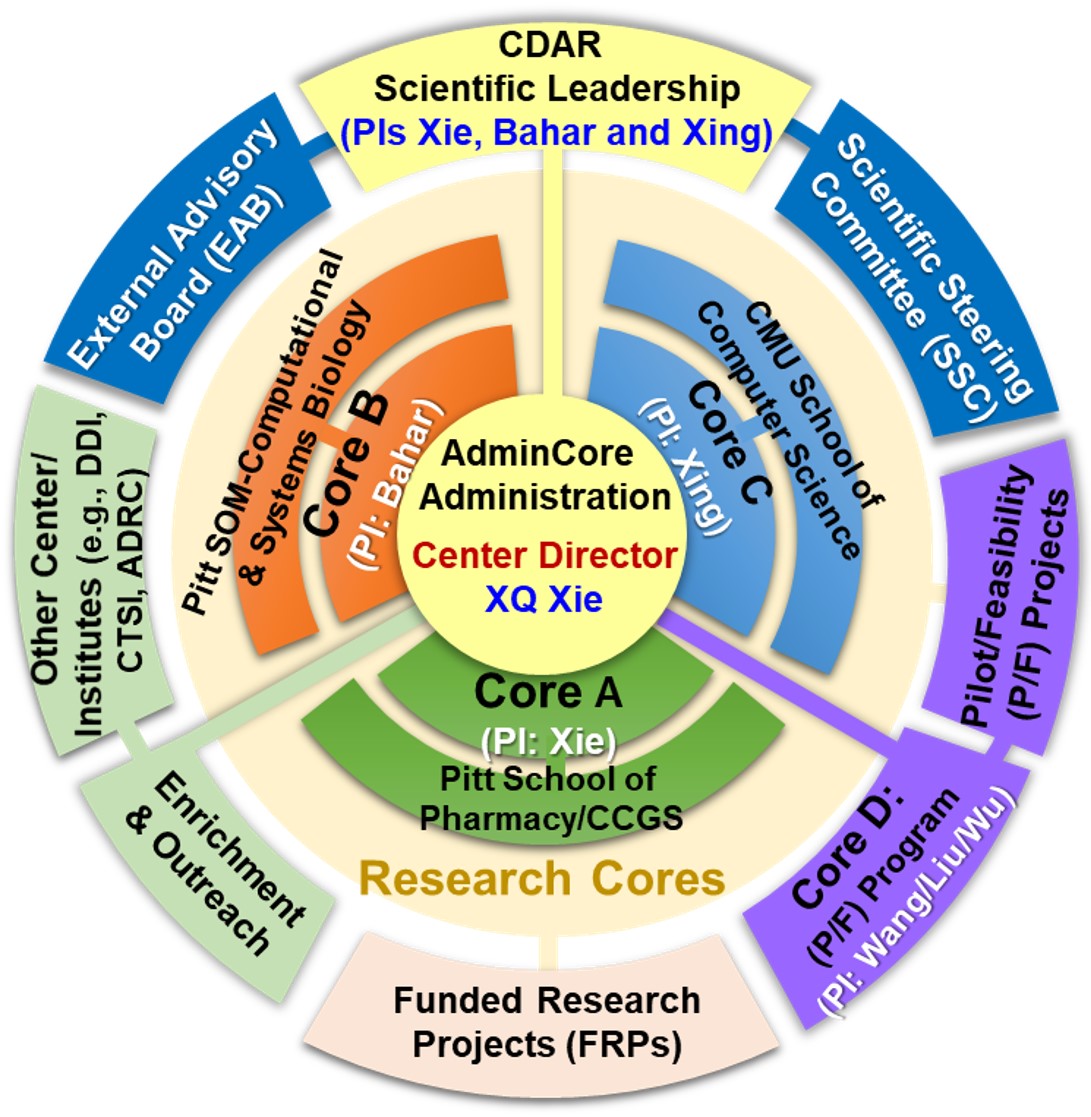
The National Center of Excellence for Computational Drug Abuse Research (CDAR), is a joint initiative between the University of Pittsburgh (Pitt) and Carnegie Mellon University (CMU), funded by the NIH National Institute on Drug Abuse (NIDA).
The goal of the NIDA CDAR will be:
- to advance and implement state-of-the-art computational chemical genomics (chemogenomics) technologies for facilitating drug abuse (DA) prevention and treatment research.
- to centralize the newly developed drug abuse chemogenomics knowledgebase (DA-KB) through a cloud computing/sourcing server platform in order to enable efficient information exchange among DA researchers and related scientific communities, and to accelerate the development of novel intervention methods for preventing and treating DA and addiction.
- to enhance DA research (DAR) by synergistically leveraging the activities of ongoing funded research projects (FRPs) and by mentoring/training junior researchers in the field.